Research A new algorithm to study millions of receptors that T cells use to recognise pathogens

Scientists from SATVI and Stanford Medicine University have developed a new method to compare and find commonalities within millions of T cell receptors (TCR), the receptors used by T cells to recognise pathogens.
CD4 T cells, also called helper T cells, act as orchestrators of the many types of immune cells that make up the “orchestra” of a healthy immune response. These helper T cells perform essential functions such as informing macrophages to become activated, making them better at killing infections. They also stimulate B cells to mature and secrete antibodies, and support cytotoxic “killer” CD8 T cells and phagocytes (cells that eat invading organisms) to protect against infections. Depletion of CD4 T cells in untreated HIV infection leads to high susceptibility to infections, such as Mycobacterium tuberculosis, in areas where both are endemic. Despite the key role played by T cells, efforts to develop a comprehensive analysis of the TCR specificities (thus revealing which pathogens these cells can recognise) have been challenging because of the diversity of human leukocyte antigen (HLA, which determine tissue type), the complexity of pathogen genomes and the limits of current methods to screen them.
"Previous methods to compare and find commonalities within TCRs lost their accuracy and efficiency when more than 10 000 TCRs were analysed.
Professor Tom Scriba.
According to Professor Tom Scriba, co-author of the paper, previous methods to compare and find commonalities within TCRs lost their accuracy and efficiency when more than 10 000 TCRs were analysed.
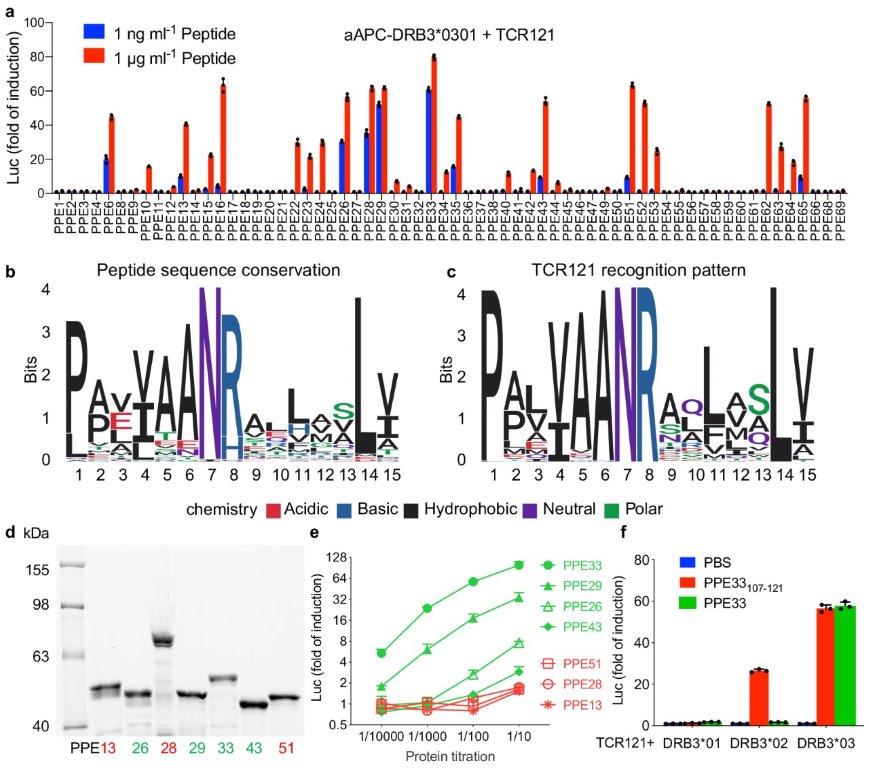
With this important advance, the new GLIPH2 method can analyse millions of TCRs, allowing enormous datasets to be studied. This creates the ability for scientists to identify TCR variants that share specificities across multiple individuals, markedly improving our abilities to study the immune response to pathogens such as the tuberculosis bacillus.
In this collaboration with colleagues from Stanford University, the GLIPH2 computational method was used to analyse 19,044 unique TCRβ sequences from 58 individuals with latent Mycobacterium tuberculosis (Mtb) infection and grouped according to their specificity.
This method was able to cluster (find groups) T cell receptor sequences according to their pathogen specificity and their ability to recognise and mediate T cell responses to small parts of Mtb proteins. The authors went further to identify the exact regions within each protein that the TCR clusters can recognise, illustrating how this technology can be used to identify successful and unsuccessful T cell responses, by comparing T cells from humans who control the bacterial infection with humans who develop TB disease. This strategy could also be used to study other pathogens and the CD4+ T cells by which they are recognised.
