Research: Differential DNA methylation of potassium channel KCa3.1 and immune signalling pathways is associated with infant immune responses following BCG vaccination

SATVI Associate Professor Tom Scriba has co-authored a research paper titled "Differential DNA methylation of potassium channel KCa3.1 and immune signalling pathways is associated with infant immune responses following BCG vaccination" appearing in the Scientific Reports journal.
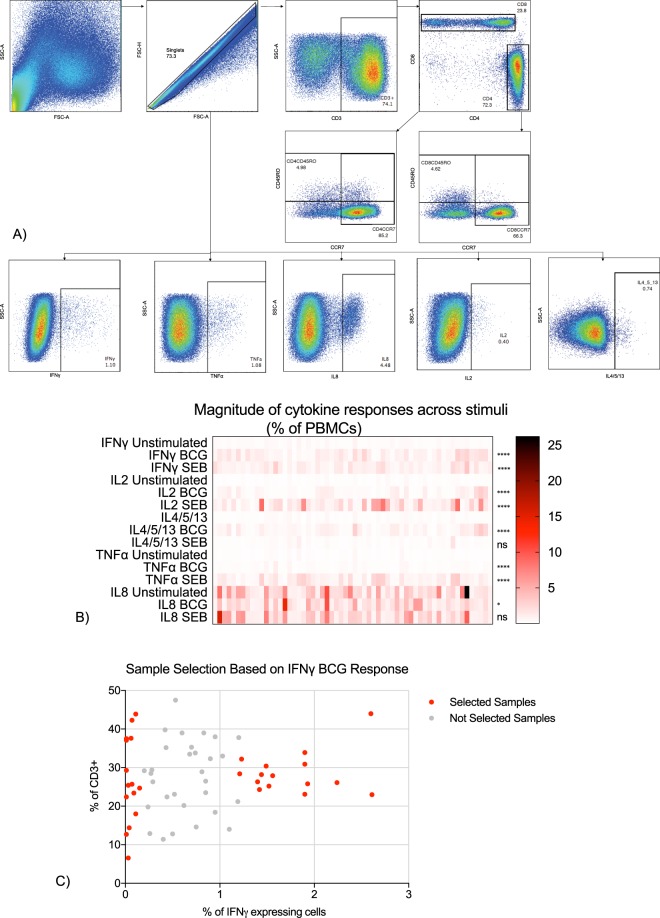
Bacillus Calmette-Guérin (BCG) is the only licensed vaccine for tuberculosis (TB) and induces highly variable protection against pulmonary disease in different countries. We hypothesised that DNA methylation is one of the molecular mechanisms driving variability in BCG-induced immune responses. DNA methylation in peripheral blood mononuclear cells (PBMC) from BCG vaccinated infants was measured and comparisons made between low and high BCG-specific cytokine responders.
Study participants
We retrieved blood samples collected from a subset of 10 week old infants who were enrolled into a large study of BCG vaccination at the South African Vaccine Initiative (SATVI) field site in the Worcester area, near Cape Town, South Africa (Hawkridge et al., BMJ 200818. Parents All participants were vaccinated with BCG at birth and samples were collected 10 weeks after birth. Participants were excluded from the study if any of the exclusion criteria were met: infant not immunised with BCG within 24 hours; mother infected with human immunodeficiency virus (HIV); chronic and acute disease in the infant at the time of enrolment; clinical anaemia in the infant; significant perinatal complications in the infant; and contact with any person with TB disease or anyone who was coughing. Participants were followed for two years to observe the development of TB disease. Only healthy infants who did not develop TB disease were selected for this study. Parents gave consent for their infants to participate in the study. The study was conducted according to the U.S. Department of Health and Human Services and Good Clinical Practice guidelines, and included protocol approval by the University of Cape Town Research Ethics Committee and written informed consent from the parent or legal guardian. The study also received ethical approval from the Ethics Committee of the London School of Hygiene and Tropical Medicine (LSHTM, #8720).
Table 1
A summary of all genes that were differentially methylated across all low and high cytokine groups.
| Name of Cytokine Pairs | Total Number of Differentially Methylated Genes | Name of Genes |
|---|---|---|
| IFNγ IL2 IL4/5/13 IL8 TNFα | 1 | ZFP57 |
| IFNγ IL2 IL4/5/13 IL8 | 2 | PTPRN2 KCNN4 |
| IFNγ IL2 IL4/5/13 TNFα | 4 | ERICH1 GPR124 RP5-1029F21.3 COLEC11 |
| IL2 IL4/5/13 IL8 TNFα | 2 | CTC-281F24.1 MIR4520-1 |
| IFNγ IL2 IL4/5/13 | 5 | RP11-609L3.1 ZMIZ1 AURKC NEURL1B SEPT7P3 |
| IFNγ IL2 TNFα | 1 | DNAH1 |
| IFNγ IL4/5/13 IL8 | 1 | AZU1 |
| IFNγ IL4/5/13 TNFα | 3 | LMTK3 LY6K ANO8 |
| IL2 IL8 TNFα | 1 | CRYGEP |
| IL2 IL4/5/13 TNFα | 1 | DTX1 |
| IL4/5/13 IL8 TNFα | 2 | EHBP1L1 SIPA1 |
| IFNγ IL2 | 2 | PRSS36 MVB12B |
| IFNγ IL8 | 1 | GCSAML |
| IFNγ IL4/5/13 | 17 | TTN DEAF1 CARS2 RP11-108K14.12 HELZ2 RGS3 COL1A1 MTHFR GRK1 DUSP15 CYP2E1 FIP1L1 LPIN1 TTN-AS1 C1orf167 FBXO41 CMTM7 |
| IFNγ TNFα | 1 | LINC00539 |
| IL2 IL8 | 1 | HCN2 |
| IL2 IL4/5/13 | 3 | PDIA3P2 CATSPER2 KCNAB1 |
| IL2 TNFα | 2 | PROSER3 MAN1C1 |
| IL8 TNFα | 3 | CUX1 PLEC SLC43A2 |
| IL4/5/13 TNFα | 9 | PKNOX2 HNRNPH1 AC010907.2 RP11-687M24.8 MAST4 LHX6 ADARB2 AC096649.3 NLGN2 |
| IFNγ | 13 | MED15P5 NTM CTD-3080P12.3 PIAS4 SPG7 TIMP2 CAMSAP3 SOGA3 CD9 MALRD1 PXDN NTNG2 RP11-599J14.2 |
| IL2 | 15 | RP4-724E13.2 TGFBR3 RP11-452L6.8 COX6A2 CARD11 NINJ2 SNRPN MAP1LC3BP1 RP11-218M22.1 C20orf96 ANK1 FANCC MEGF6 RP4-559A3.7 LEFTY1 |
| IL8 | 88 | RN7SL646P FAM222B ZFP41 PBX4 LINC01169 KCNQ1OT1 TRPV2 EDARADD UMODL1-AS1 DTNB bP-21264C1.2 RP11-216L13.18 KIAA1161 CDH23 ZNF473 LINC01266 COL23A1 AC011850.2 FLJ26850 RP11-394I13.3 LINC00467 SOX10 MSL3P1 AC018688.1 HIST1H3E FAM217B 5_8S_rRNA RNMTL1 RAI1 RELT LRRC24 NFATC1 UNC93B1 ZNF790-AS1 EMC9 ITPR1 POU5F1 EGOT UMODL1 MIR4508 AC005481.5 VOPP1 AC005538.3 ATP8B3 POLR2F MIR6820 SDK1 MIR138-2 FRRS1 MIR6813 WDR60 COL5A1 CTD-2126E3.3 THSD4 OPCML TMPRSS12 RABL6 RP11-133L14.5 PROSER2-AS1 PIK3R2 PRSS21 RYR1 CLEC9A CTD-2269E23.4 MAN2B1 ARHGEF10 GLI4 RP11-497G19.1 ATG16L2 LZTS3 WDR20 SH3GL1 EXD3 AMOTL2 LRRC14 MROH1 CTD-2192J16.22 TARID CFD TNFRSF11A MMP17 RGS19 KCNQ1 C8orf82 RNF165 LOXL1 LINC01312 SYCP2 |
| IL4/5/13 | 41 | TPM4P1 GNAL AC131097.4 RP11-244F12.1 RP11-867G23.12 NBPF13P RNVU1-8 RIMBP2 LINC01237 ZNF718 RP11-680G24.4 TAF1C TBCE PHACTR3 RP11-662M24.2 RP11-452D2.2 RP11-545A16.3 HLA-DRB9 PRDM15 KCNN3 PRDM16 DRAXIN ARID3A SNRNP27 COL18A1 RP11-1260E13.1 RPH3AL RP3-522J7.6 AP001347.6 LRRC37A6P PRDM7 MYBPC2 PNPLA6 AC147651.1 PDXDC1 SRC NYAP2 ADAMTS12 ANKRD20A18P C5orf17 RIN1 |
| TNFα | 98 | GABRB3 MKRN3 PRKAR1B PAX8-AS1 FAM189A1 RNA5-8S5 ACOT2 WT1 ACVR1B LINC00839 AOX3P PRDM8 LDLRAD2 CHCHD3 FBXO16 PEBP4 MARK4 CTU1 HLA-DPA1 ATP5J2 RPL13A MEST RASAL2 PRAM1 PRKCQ GNAI2 RP11-127L20.3 RP11-726G23.10 BRSK1 BCAN CROCC GABRA5 LINC00887 PTCD1 SDF4 FNDC3B RPL13P12 CYGB NCOR2 CLIP2 GALNT9 BCR AOX2P OPRL1 RP11-377G16.2 FLT3LG CTD-3148I10.9 CPT1A RP11-54O7.17 RPS6KA4 RP11-284F21.7 PIGQ RP11-492E3.2 PRCD AC093609.1 HLA-DPB1 SEMA6B CH507-513H4.1 UHRF1 CTD-3148I10.15 SEMA3B ZNF331 GEMIN7 MPPED1 ERICH1-AS1 RP5-906A24.1 PPAPDC3 MGRN1 ZNF395 FLT4 ITPRIP RP11-426C22.5 KAZN MST1L PSPN ITFG2 ARL5C RP11-137H2.4 FBXW4P1 PAX8 ALKBH7 C17orf98 RBM19 NOS3 CCNYL2 RP11-273G15.2 HTRA4 RP11-231D20.2 RP11-65I12.1 SCARF1 ITGAX RP11-230C9.1 PPP1R37 RP11-56P9.10 OSBPL5 ATP5J2-PTCD1 CBFA2T3 PLEKHA2 |
Each row shows single or multiple genes that were differentially methylated across one or multiple cytokine groups. n = 30.
We found 318 genes and 67 pathways with distinct patterns of DNA methylation, including immune pathways, e.g. for T cell activation, that are known to directly affect immune responses. We also highlight signalling pathways that could indirectly affect the BCG-induced immune response: potassium and calcium channel, muscarinic acetylcholine receptor, G Protein coupled receptor (GPCR), glutamate signalling and WNT pathways.
This study suggests that in addition to immune pathways, cellular processes drive vaccine-induced immune responses. Our results highlight mechanisms that require consideration when designing new TB vaccines.
