Research: Differential Recognition of Mtb-specific Epitopes as a Function of Tuberculosis Disease History

SATVI authors Professor Mark Hatherill, Associate Professor Tom Scriba, Drs. Munyaradzi Musvosvi and Virginie Rozot have co-authored a research paper titled "Differential recognition of Mtb-specific epitopes as a function of tuberculosis disease history" appearing in the American Journal of Respiratory and Critical Care Medicine.

Abstract
Rationale: Individuals with a history of tuberculosis (TB) disease are at elevated risk of disease recurrence. The underlying cause is not known, but one explanation is that previous disease results in less-effective immunity against Mycobacterium tuberculosis (Mtb).
We hypothesized that the repertoire of Mtb-derived epitopes recognized by T cells from individuals with latent Mtb infection differs as a function of previous diagnosis of active TB disease.
Objectives: We hypothesized that the repertoire of Mtb-derived epitopes recognized by T cells from individuals with latent Mtb infection differs as a function of previous diagnosis of active TB disease.
Methods: T-cell responses to peptide pools in samples collected from an adult screening and an adolescent validation cohort were measured by IFN-γ enzyme-linked immunospot assay or intracellular cytokine staining.
Measurements and main results: We identified a set of "type 2" T-cell epitopes that were recognized at 10-fold-lower levels in Mtb-infected individuals with a history of TB disease less than 6 years ago than in those without previous TB. By contrast, "type 1" epitopes were recognized equally well in individuals with or without previous TB. The differential epitope recognition was not due to differences in HLA class II binding, memory phenotypes, or gene expression in the responding T cells. Instead, "TB disease history-sensitive" type 2 epitopes were significantly (P < 0.0001) more homologous to sequences from bacteria found in the human microbiome than type 1 epitopes.
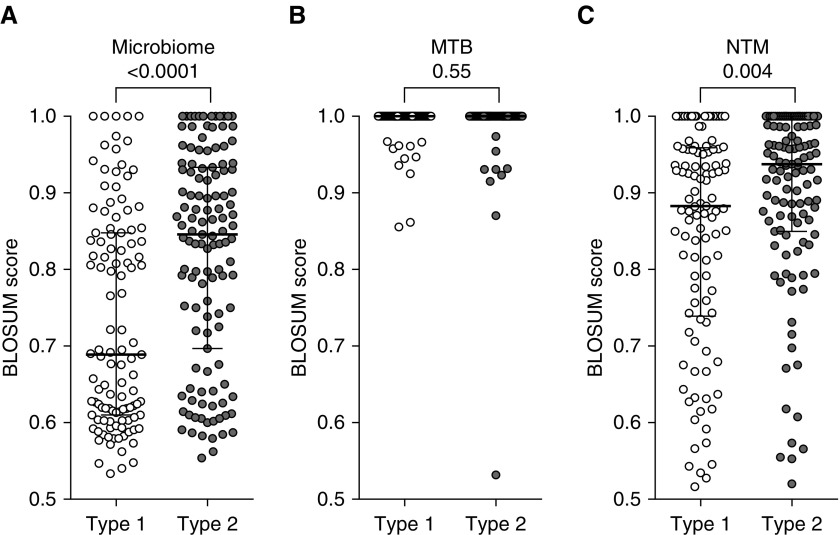
Conservation of type 1 (n = 113; white dots) and type 2 (n = 123; gray dots) epitopes in (A) the human microbiome, (B) Mtb complex strains, and (C) NTM strains is shown. Each dot represents the maximum BLOSUM score per individual peptide (i.e., the highest homology found for each peptide within the human microbiome, Mtb complex strains, or NTM strains). Median and interquartile range are indicated. The numbers above the brackets are P values determined by two-tailed Mann-Whitney test.
