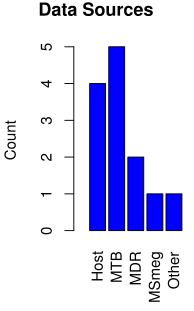
UCT has several Tuberculosis researchers tackling the problem of TB and TB-related issues. In order to be more effective in dealing with TB, collobration between groups are crucial. We have summarised the current UCT human resources involved in Mycobacterium (MTB) and MTB host data generation and analysis.
|

|
Current MTB Research Data at UCT
| Principal investigator | Data source: Type |
| Robert Wilkinson | MTB: WGS |
| Tom Scriba | Host: Whole blood RNA-Seq, Plasma proteomic |
| Digby Warner | MTB: WGS |
| Anna Coussens | MDR MTB: WGS |
| Valerie Mizrahi | MTB: Tn-seq; MTB: Tn-seq |
| Keertan Dheda | Host: BAL RNAseq, lung tissue RNAseq, urine proteomics; MTB: WGS, RNAseq, proteomics; MDR and XDR TB: WGS and RNAseq |
Several tools exist for investigating TB drug resistance mutations in sequence data:
TB Profiler: Given raw sequence data as input, TB Profiler infers strain type and identifies known drug resistance markers. This in silico diagnostic uses a library generated from 1,325 mutations predictive of drug resistance for 15 anti-tuberculosis drugs. The library was validated for 11 of these drugs using genomic-phenotypic data from 792 strains (Coll et al).
ReSeqTB: "...catalogs a vast amount of genotypic, phenotypic and related metadata from Mycobacterium tuberculosis (Mtb) strains to enable the development of clinically useful, WHO-endorsed in vitro diagnostic assays for rapid drug susceptibility testing of MTb..."
MtbClade Identifier: "...fast identification of known clades/genotypes of Mycobacterium tuberculosis by complete genome sequences, sequences of predicted genes and aligned NGS reads..."
A good recent (August 2016) review and summary of the status of computational pan-genomics have been compiled by The Computational Pan-Genomics Consortium.
