|
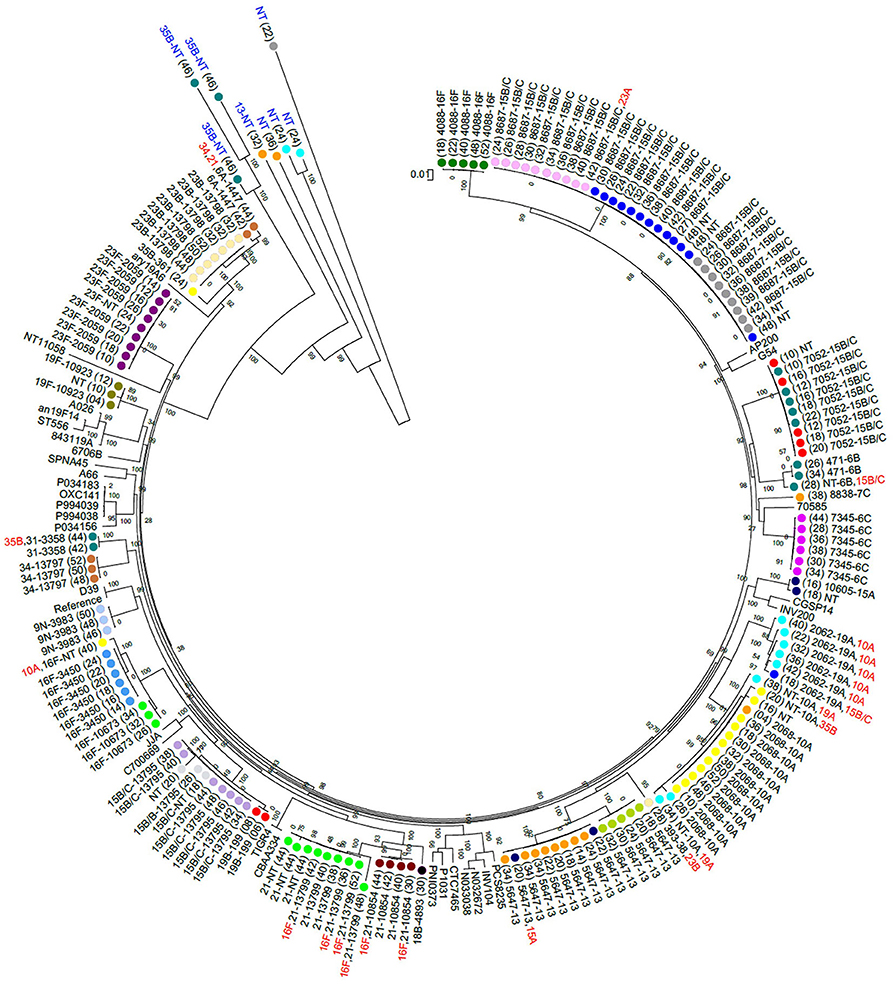
| Figure 1: A maximum likelihood whole genome phylogenetic tree of pneumococcal isolates recovered from 23 South African infants |
|
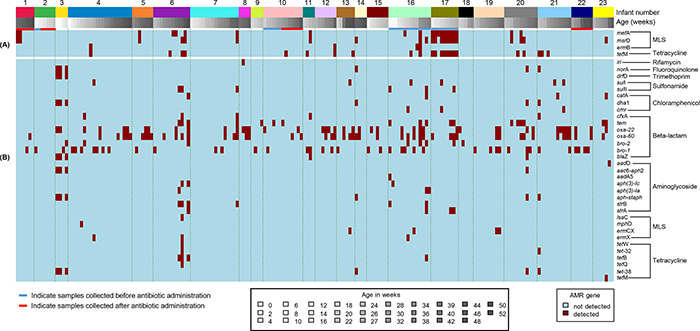
| Figure 2: Shotgun metagenomic sequence-derived molecular sequence type and resistome (from direct sequencing of NP swabs), and phenotypic antimicrobial susceptibility results (from pneumococcal isolates) obtained from 23 infants (assigned 1 to 23, top row) |
| Nasopharyngeal colonization with antimicrobial-resistant bacteria is a global public health concern, and antimicrobial-resistance genes carried by the resident microbiota may serve as a reservoir for transfer of resistance elements to opportunistic pathogens. In a study investigating pneumococcal colonization dynamics and antimicrobial resistance in South African infants over the first 2 years of life, shotgun metagenomic sequencing of nasopharyngeal samples was able to fully characterise longitudinal changes as different sequence types colonized this niche. in silico analyses defined the genetic relatedness (Figure 1), and described the transient resistome as resistance determinants were gained or lost, as strains were replaced over time (Figure 2). These novel findings indicated a complex interplay between bacteria colonizing the nasopharynx from birth, and highlighted the important role of next generation sequencing technology in culture-independent microbial research. |
| Links to publications: https://doi.org/10.3389/fpubh.2020.543898 https://doi.org/10.1371/journal.pone.0231887 https://doi.org/10.3389/fmicb.2019.00610) |