Research in our lab focusses on understanding the dynamics of gene regulation in the nervous system during cell development, maturation and infection, as well as exploring the evolution of these processes. We combine genome-wide analyses of gene expression and chromatin conformation to identify genes and cis-regulatory elements that are being dynamically modified as cells develop, mature or respond to infection. We use in vivo and in vitro systems to verify gene expression profiles and gene regulatory activity in embryos and tissue explants. There are three major research projects in the lab:
1. The evolution of gene regulation during neural crest development
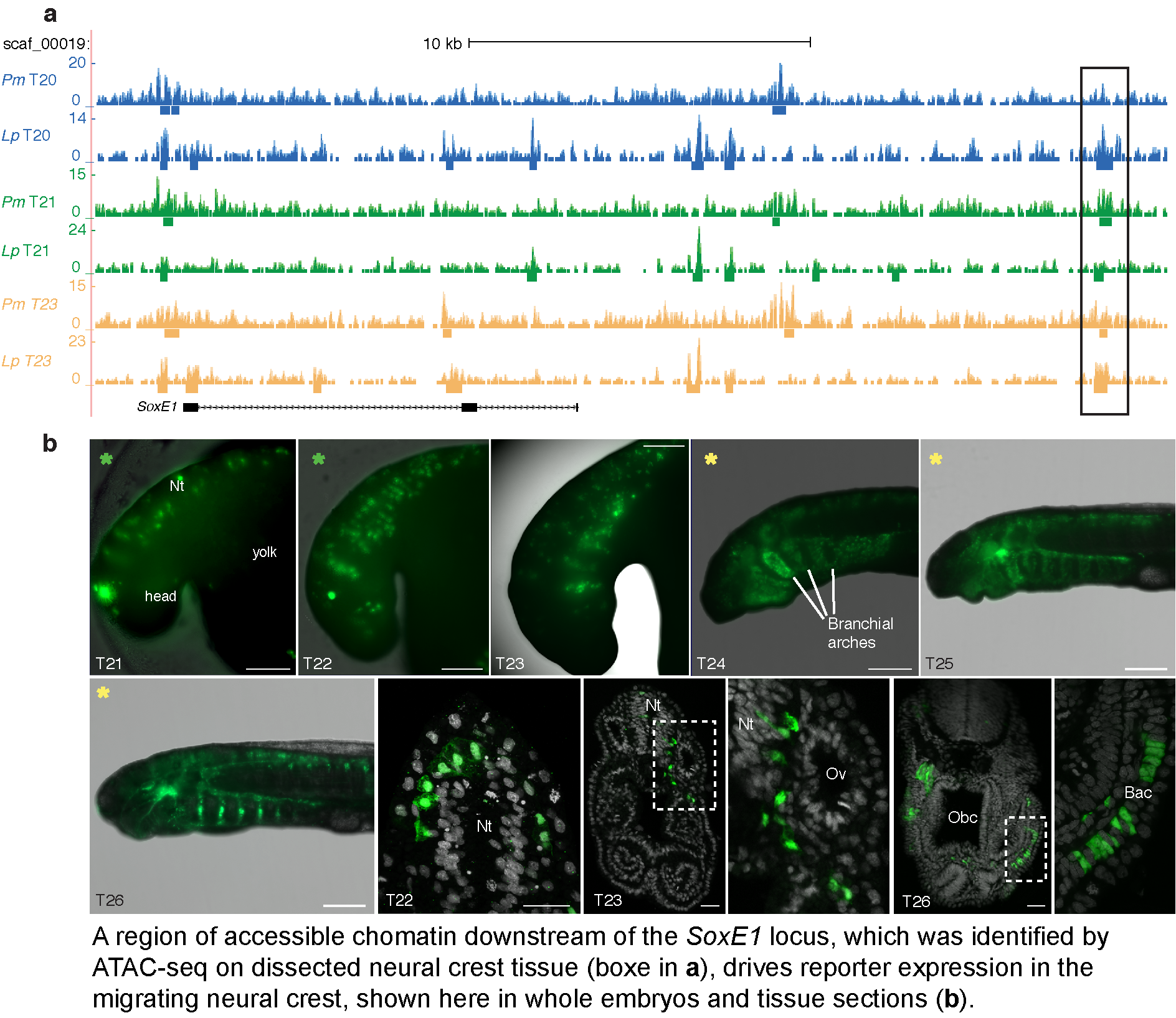
This project uses the lamprey, the most basal vertebrate, as a model to determine the genetic programme for one of the defining features of the vertebrates: the neural crest. The neural crest is a migratory embryonic cell population that contributes to many critical adult features including the skull, teeth and peripheral nervous system. Knowledge of the “recipe” for making neural crest cells will help us determine the causes of neural crest defects in humans, and will also assist in harnessing the potential of this tissue for therapeutic purposes. While the neural crest has been studied extensively we do not know how it was first specified in our evolutionary ancestors. We are tackling this problem using the latest genome-wide analysis techniques, including bulk RNA-seq, single cell RNA-seq and ATAC-seq, in the lamprey for the first time. By comparing the results to similar datasets for other vertebrates, such as chicken and zebrafish, we are uncovering critical regulatory elements and sets of expressed genes that are shared across vertebrates and others that are either present in lamprey alone or missing from the lamprey. In this way, this research focuses attention on the regions of the genome that gave neural crest cells their properties in the earliest vertebrate ancestors and form the core recipe for specifying this unique cell type in modern vertebrates.
2. Gene regulatory dynamics of human brain maturation
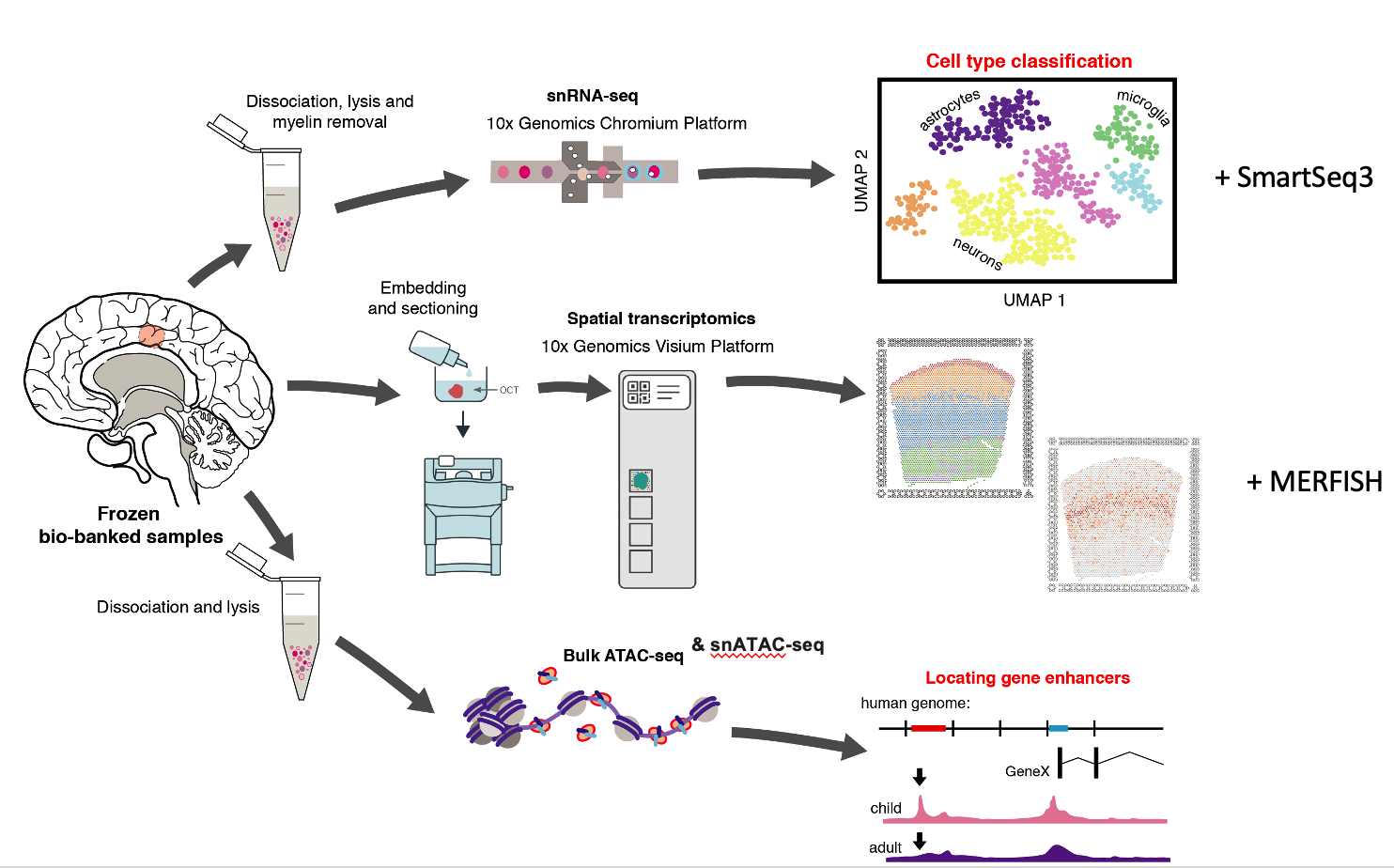
The human brain is a complex assembly of diverse cell types with distinct morphologies and functions. Each cell type has a distinct molecular profile with tightly controlled gene regulatory networks likely in play directing cell differentiation and maintaining correct cell function. This molecular signature is set up over a protracted period of development, which begins in the embryo and continues into adolescence. Is it likely that over the course of a human lifetime, the molecular profile of the mature brain continues to change in response to environmental impacts and the process of ageing. However, due to the scarcity of healthy ante-mortem brain tissue samples, very little is known about how the normal gene expression and gene regulation profiles of the human brain change as it matures, or how this molecular signature is altered in response to environmental challenges, such as infectious disease. Through collaborations with local neurosurgeons we are seeking to address these short-falls in our understanding of the human brain. Using a biobank of human brain tissue obtained during surgery, which encompasses a wide range of ages, we are using single nucleus RNA-seq and ATAC-seq to describe and track the cell-type specific gene regulatory dynamics that occur as the brain matures. Overall, this study will provide a baseline view of the gene regulatory network dynamics that are play as the human brain matures from childhood to adulthood. By focusing on samples collected from a local population , these datasets will form an invaluable resources to local biomedical researchers and medical practitioners, looking to understand how diseases such as TB and HIV impact on the normal functioning of the human brain.
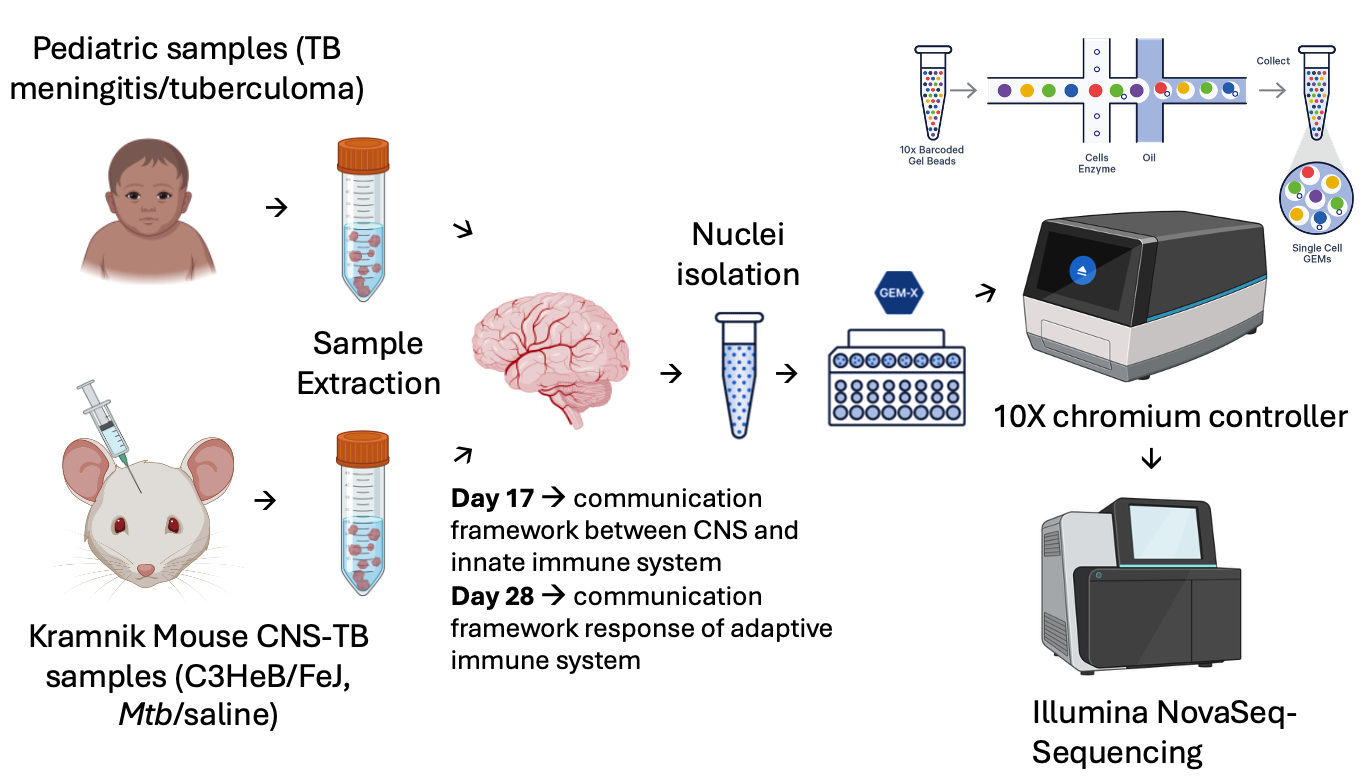
3. Cell-type-specific responses to central nervous system tuberculosis (CNS-TB)
CNS-TB is the most severe extra pulmonary form of the tuberculosis, which can have a fatality rate of up to 80%. Significant barriers to understanding disease pathogenesis exist, which translates to poor diagnosis and treatment outcomes. Improved insight into the cell-type-specific responses to infection in CNS-TB will help us understand the mechanisms associated with the development of CNS-TB and different CNS-TB outcomes. Ultimately this will allow us to develop novel, effective, and broadly accessible therapeutics. In collaboration the African Brain Child group at the Neuroscience Institute and the Experimental Tuberculosis and Immunology Research group in the Institute of Infectious Disease & Molecular Medicine, we are using single cell RNA-seq and spatial transcriptomics approaches to investigate the transcriptional changes induced by M. tuberculosis infection of the brain under clinical and experimental conditions.