At COIN, DNA sequencing lies at the heart of how we uncover the genetic causes of neurological and rare disorders. By reading the sequence of DNA bases that make up the human genome, we can detect variants that disrupt normal biological function and ultimately, help explain disease. Our team combines multiple sequencing technologies, each with unique strengths, to build a complete picture of genetic variation across diverse African populations. We use these tools strategically, balancing scientific depth with cost-efficiency, to ensure that precision medicine remains accessible and relevant in a resource-limited setting.
Next Generation Sequencing (NGS): Speed and Scale
Next Generation Sequencing (NGS) refers to modern high-throughput methods that allow millions of DNA fragments to be sequenced simultaneously. Compared to older, one-gene-at-a-time approaches, NGS offers a faster, more affordable way to explore large regions of the genome at once. At COIN, NGS forms the backbone of our diagnostic pipeline. It enables us to detect single-nucleotide variants, small insertions and deletions and structural changes that contribute to neurological diseases. By integrating NGS data with ancestry -aware analyses, we can also identify population-specific patterns of genetic variation, a key step toward understanding Africa’s rich genomic diversity.
Why this Work Matters
Short-Read Sequencing: Precision and Affordability
Short-read sequencing, often performed using Illumina platforms, is the most widely used form of NGS. It generates millions of short fragments (typically 100–300 base pairs long) that can be aligned to a reference genome or pangenome for variant discovery. Its main advantages are accuracy, throughput and cost-effectiveness, making it a practical choice for both research and clinical diagnostics. At COIN, we use short-read sequencing for whole-exome sequencing (WES) and targeted gene panels to efficiently identify variants in known disease genes. In settings where resources are limited, short-read data allow us to maximise diagnostic yield at an accessible cost, while applying machine learning and bioinformatics tools to overcome technical limitations such as incomplete coverage or copy-number uncertainty.
Long-Read Sequencing: Seeing the Bigger Picture
Long-read sequencing, offered by platforms such as PacBio , can read DNA fragments thousands of bases long. This provides a much clearer view of complex genomic regions, repetitive sequences , and large structural variants that short reads may miss. Although currently more expensive and less scalable than short-read methods, long-read sequencing offers critical advantages, particularly for discovering novel variants and resolving regions of the genome that are difficult to map. At COIN, we integrate long-read data to validate complex variants, phase alleles, and refine findings from our exome and short-read analyses. This hybrid approach allows us to achieve the accuracy of high-end genomic research while keeping the overall process cost-conscious.
Whole-Exome vs Whole-Genome Sequencing: Depth and Breadth
Both Whole-Exome Sequencing (WES) and Whole-Genome Sequencing (WGS) are essential tools in our research, each serving distinct purposes:
- Whole-Exome Sequencing (WES) focuses on the protein-coding regions of the genome - about 1–2% of our DNA - where most known disease-causing mutations occur. It is affordable, efficient, and ideal for identifying variants linked to Mendelian neurological disorders.
- Whole-Genome Sequencing (WGS) reads the entire genome, capturing both coding and non-coding regions. This provides a more complete view of genetic variation, allowing for the discovery of noncoding, regulatory, or structural variants that may influence disease risk.
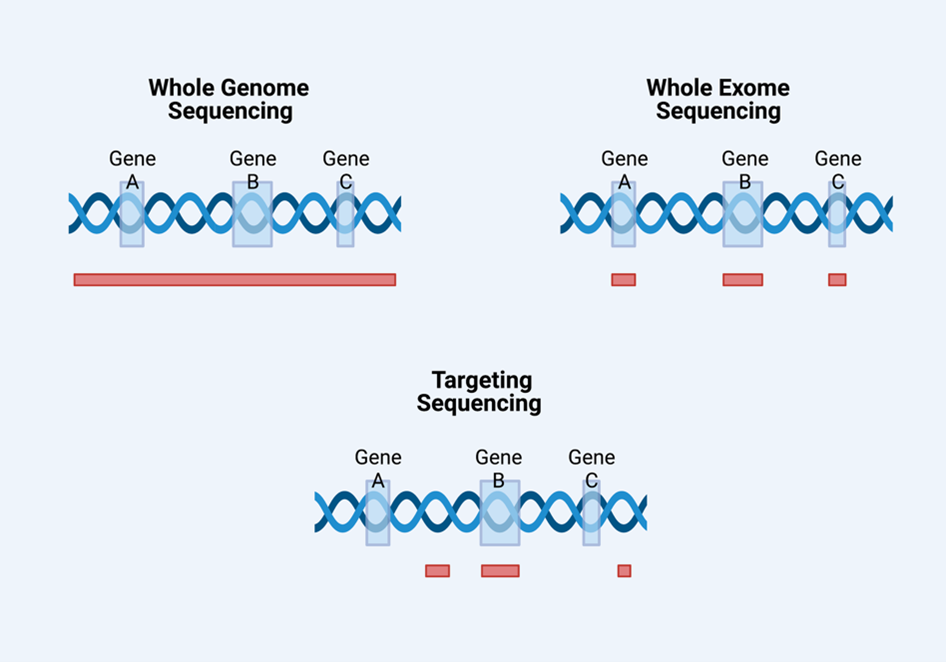
At COIN, we often begin with WES for diagnostic screening, then use WGS selectively for complex or unresolved cases. This tiered strategy maximises insight while minimising cost; an approach that aligns with our mission to make advanced genomics practical for African healthcare systems.
A Resourceful Approach to Precision Medicine
By combining short-read and long-read technologies and using both exome- and genome-level data, we are building a flexible sequencing framework suited to Africa’s unique challenges and opportunities. Our goal is to deliver high-impact genetic insights without relying solely on expensive infrastructure. Through this adaptive use of sequencing, COIN is advancing the diagnosis and understanding of neurological and neuromuscular disorders, helping ensure that the benefits of genomics are shared equitably, and that African genetic diversity is meaningfully represented in global precision medicine.





