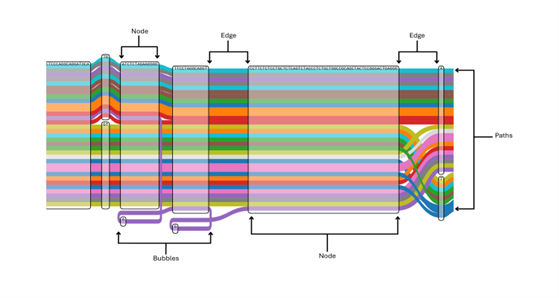
About 99% of the human genome is similar across all individuals globally. Efforts have been made to develop a human genome reference to aid in understanding variations and disease mechanisms. Albeit one single human reference genome cannot fully represent the remaining percentage difference that contribute to the massive diversity across different populations. This is where pangenome research comes in. A pangenome is a complete collection of all genes/genomes present in a particular species or population. Pangenome research fills in the gap created by linear reference because it creates a more comprehensive and diverse reference that represents all variations within a population. This comprehensive representation is based on a graph model where all multiple alternative sequences coexist. As shown in Fig 1 below, a pangenome graph consists of three main components: nodes, which represent segments of DNA; edges, which connect the nodes and indicate possible sequences; and paths, which are routes through the nodes and edges representing individual genomes or haplotypes. This approach helps capture population-specific variants, structural rearrangements and deconvolutes complex regions that are often missed by the traditional linear reference approach. Graph pangenomes can therefore, help scientists gain better understanding of the genetic architecture of rare diseases which has hitherto been difficult to explore using recent methods. Structural variants are known to contribute to some of these rare diseases including neuromuscular and neurological diseases. However, linear reference genome approaches often fail to capture these complex variants. Utilising pangenomic approaches can, therefore, deepen our understanding of human diversity and advance technological advances towards precision medicine.
Why This Research Matters
Pangenomic research seeks to make genomic medicine more inclusive, accurate and equitable. One of the main challenges pangenomes address is underrepresentation of some populations, especially Africans, in widely used reference genomes which are traditionally based on individuals of European ancestry. Recent efforts by the Human Pangenome Reference Consortium (HPRC) are beginning to address this challenge. For instance, the first human pangenome released in 2023 by the HPRC is composed of 47 phased, diploid assemblies from diverse ancestral backgrounds around the world. Of these, nearly half are of African ancestry. Including Africans, populations harbouring the greatest diversity globally, ensures that no one is left behind in the rapidly evolving landscape of genomic research. Additionally, pangenome-based approaches help improve structural variant discovery, mapping accuracy and population-level genetic diversity capturing. These advantages propel efforts in advancing precision medicine and understanding genetic architecture of rare and common diseases. In Africa, researchers at eLwazi Open Data Science Platform (ODSP) are pioneering the building of the first African-only pangenome graph (RefGraph Project) to better understand the diversity of genomes on the continent and drive translational efforts in genomic medicine.
Looking ahead
The COIN Unit is positioned to expand its efforts and contribute to the rapidly evolving pangenome field. Armed with an able team comprising bioinformaticians, clinicians and geneticists we look forward to bridging the gap between genomic discovery and clinical application.

