What are short tandem repeats (STRs)?
Tandem repeats (TRs) are DNA sequences that contain a motif of 1 or more nucleotide(s) repeated consecutively. For example, the motif ‘AT’ may be repeated several times, or ‘CTG’ repeated multiple times. Tandem repeats may be broadly categorized into short tandem repeat (STR), where the repeat unit is 1-6 bp (Figure 1A), or variable number tandem repeat (VNTR) where the repeat unit is 7 bp or more (Figure 1B).
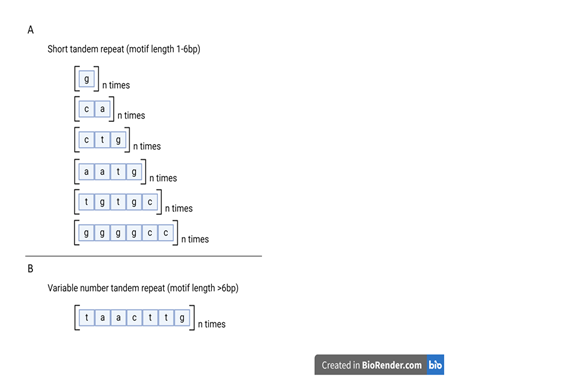
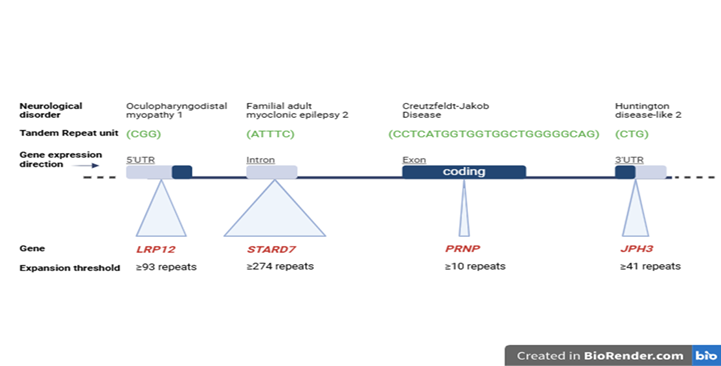
TRs are found throughout the genome, including exonic, intronic, intergenic and promoter regions. In some cases, expansions of these repeats are linked to distinct disease phenotypes due to their location within specific genomic regions (Figure 2). Beyond their role in causing neurological or neuromuscular diseases, these tandem repeats serve as biomarkers for population studies or forensic identification. The same TR motif/unit may vary in length between individuals: for instance, one person may have the motif ‘AT’ repeated four times (AT₄), while another may have it repeated seven times (AT₇). It is in these unique properties in repeat length that make TR motifs highly informative markers for genetic diversity, disease susceptibility and individual identification.
Expanding the role of STRs in Neuromuscular disease research
The genetic pathogenesis of motor neuron or neuromuscular diseases is often driven by STR expansions, with over 30 Mendelian disorders reported to be caused by the inheritance of STR expansions. At the COIN Unit, we are advancing the study of STRs using whole exome sequencing (WES), a powerful tool that captures the protein-coding regions of the genome. While many research projects that investigated WES data typically focused on single nucleotide variants (SNVs), our research expands its utility to detect and analyse STRs with high resolution and accuracy.
Some current work that we have done is to genotype STRs in neuromuscular disease as well as to build a database where we show the population-specific variation in healthy individuals so that we may clearly understand TR variation in disease. Furthermore, we are interested in repeat length variation in the control vs patient cohort group to help towards understanding the underlying mechanisms of disease. When considering longitudinal work that may evolve from diagnosing TRs linked to disease, gene therapies targeting these specific TR expansions in patients may then be developed, presenting a more viable approach to patient care.
Towards more comprehensive repeat detection
The current analysis pipeline for investigating TRs in the COIN unit is to align genomic sequences to the GRCh38 reference genome and then use specialized tools (such as ExpansionHunter or STRipy) to genotype STRs. Using the pangenome as a reference seems to be useful to identify more STRs that might be unique and therefore not properly represented in the GRCh38 reference genome. Current STR genotyping tools rely on the reference genome to call TRs/STRs in individuals, and so use of the pangenome might be helpful to this end. Moreover, we are eager to explore long-read sequencing and thereafter, generate TRGT (Tandem Repeat Genotyping Tool) reports that leverage extended read lengths to resolve previously unsolvable expanded STRs as well as VNTRs, which are too large for short-read sequencing.